Introduction
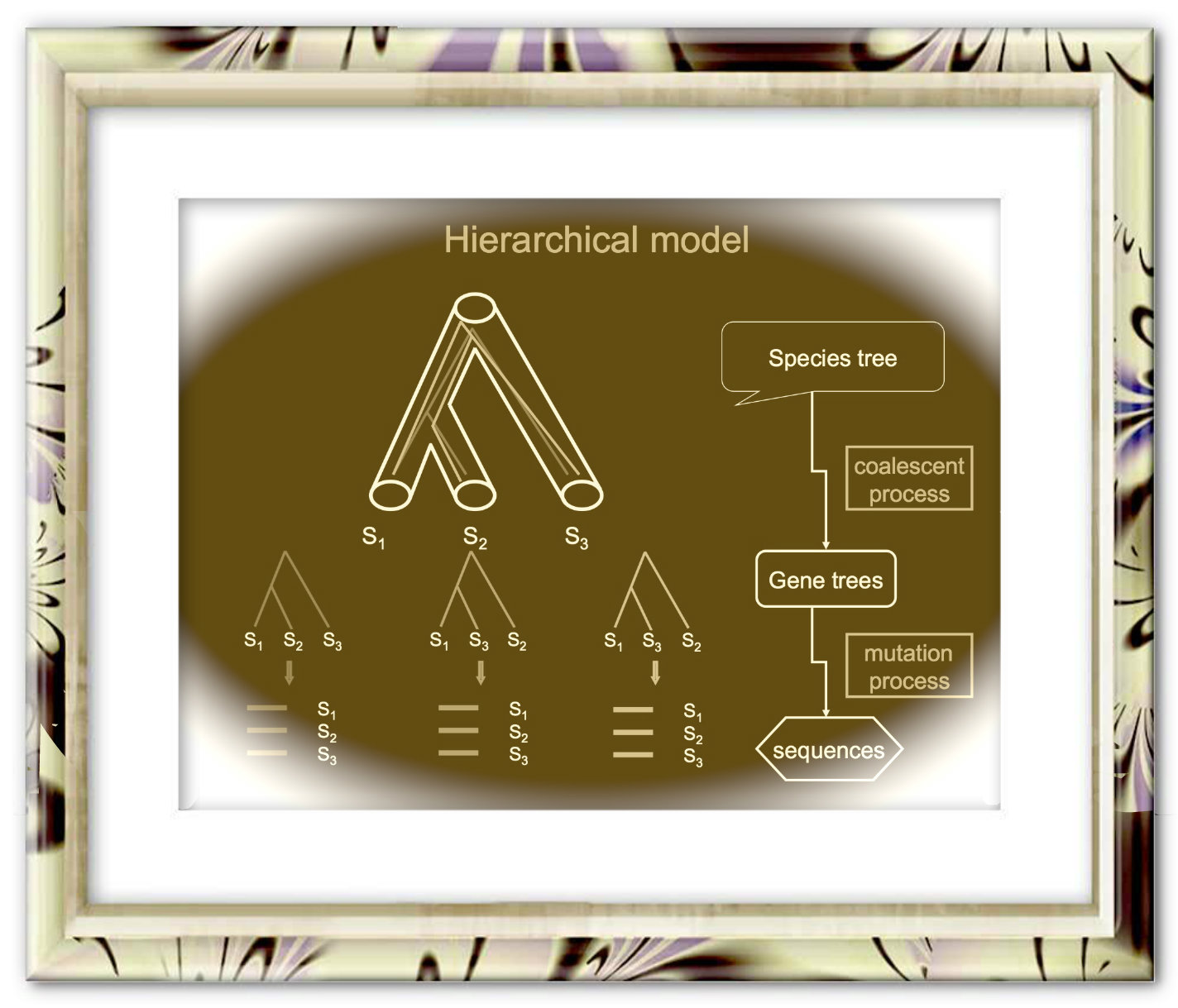 The species tree reconstruction webserver [1] consists of a series of phylogenetic tools for estimating species trees from phylogenomic data: 1) rooting gene trees with outgroup species, 2) estimating species trees from the rooted gene trees using STAR [2], MP-EST [3], NJst methods [4], 3) bootstrap analyses for the species tree reconstruction methods (STAR, MP-EST, and NJst).
The species tree reconstruction webserver [1] consists of a series of phylogenetic tools for estimating species trees from phylogenomic data: 1) rooting gene trees with outgroup species, 2) estimating species trees from the rooted gene trees using STAR [2], MP-EST [3], NJst methods [4], 3) bootstrap analyses for the species tree reconstruction methods (STAR, MP-EST, and NJst). *Note* We've removed the cluster version back to development due to recurring disconnectivity with UGA GACRC zcluster. As a result, we added additional job processing power within our server to run multiple job submissions.
News
October 20 2021-- Moved the website to straw.phylolab.net April 5 2021-- Updated MP-EST to the latest version MPEST2.0.
March 13 2013-- Added capacity for running multiple jobs at the same time. (Previously could only process one job at a time);
March 14 2013-- Fixed problem with embedded tree files within bootstrapping.
March 17 2013-- Added function for taking input trees with inconsistent branch length.
March 19 2013-- Added recogniztion of *.nwk as file type for bootstrap zipped file. Previously only files ending with file type *.tre, *.phylip, *.txt and *.new were accepted for the zipped file for bootstrapping.
April 03 2013-- I talked with some of the user and realized that some users found their jobs still running. There was a power outage and affected the running jobs.
April 03 2013-- Added report style distance measures.
April 03 2013-- Added species tree branch distance within bootstrap tre file result.
June 13 2013-- There were some hardware issues with the server and we had to reboot the system. Sorry if any jobs were submitted that showed invalid input etc... The pipeline stopped working due to some hardware problem.
Dec 11 2014-- Updated the bootstrapping feature to lower the memory usage.
References
[1] Shaw, T., Z. Ruan, T. Glenn, and Liu, L. STRAW: Species TRee Analysis Web server. Nucleic Acids Research. 2013, doi: 10.1093/nar/gkt377
[2] Liu, L., L. Yu, D.K. Pearl, and S.V. Edwards. Syst. Biol. 2009, 58(5):468-477.
[3] Liu, L., L. Yu, S.V. Edwards. BMC Evol. Biol. 2010, 10:302.
[4] Liu, L., and L. Yu. Syst. Biol. 2011, 60: 661-667.
Contributors
Timothy Shaw, Zheng Ruan, Travis Glenn, Liang Liu.
(Website designed by Albert Shaw
For questions, contact lliu@uga.edu.
Last Updated Date: Oct 20, 2021
Sponsored By


Disclaimer
Please use a modern browser (ie Firefox, Safari, Chrome) with javascript enabled to have the best viewing capability of the result from the species tree analysis. At this time, the regular web server cannot perform species tree analyses involving more than 200 gene trees or greather than 50 species. We are in the process of developing a cluster version of the server for handling larger queries, or you may download the software and run the analysis on your local computer. If you need help with running large queries send an email to lliu@uga.edu.
 STRAW: Species TRee Analysis Web server
STRAW: Species TRee Analysis Web server